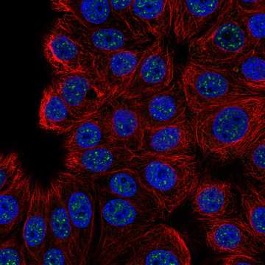
Figure 1: A fluorescent microscopic image of the localization of the protein C16orf71 to areas in cells called ‘nuclear speckles,’ colored green in this picture. Nuclear speckles are responsible for storing RNA splicing factors in cell nuclei, and proteins like SRRM2 and SON are responsible for creating the structure of these speckles
Source: Wikimedia Commons
As a key part of all eukaryotic cells, the nucleus plays an important role in storing a cell’s DNA and serving as the locale for its transcription into RNA, which provides the template for protein synthesis in the cytoplasm. One of the more interesting, albeit mysterious, parts of the nucleus are the “nuclear speckles,” otherwise known as ‘interchromatin granule clusters.’ First observed by physician Santiago Ramon y Cajal in 1910, these speckles contain high concentrations of transcription and RNA splicing factors (Lamond and Spector, 2011). These splicing factors are responsible for removing “introns,” sections of RNA that do not code for proteins, thereby playing a key role in processing the initial RNA strand into one ready for translation (Clancy, 2008). Nuclear speckles are believed to be responsible for regulating the presence of nucleic splicing factors, and have been implicated in the spread of Influenza A and the Herpes simplex viruses. (Spector and Lamond, 2003; Ilik et. al, 2020).
Recent work from Ilik et. al has revealed two proteins, named ‘SON’ and ‘SRRM2’ that may be responsible for the structural development of nuclear speckles. In an attempt to search for these molecules, the group decided to use the monoclonal antibody SC35 (Ilik et. al, 2020). SC35 is normally used to identify and stain nuclear speckles and works by binding to SRSF2, a splicing factor protein often associated with speckles (Fu and Maniantis, 1990). The group conducted an immunoprecipitation mass spectrometry experiment in which the binding targets of SC35 were precipitated from solution, concentrated, and characterized via mass spectrometry – a process where a molecule is ionized and then deflected through a magnetic field to measure the mass of its component ions (Ilik et. al, 2020). To their surprise, the group found that SRRM2 had the highest concentration in the immunoprecipitation-enriched sample of the binding targets of SC35, suggesting that SRRM2 is its primary target. This was further confirmed through immunoblot and immunofluorescence staining (Ilik et. al, 2020).
Ilik et. al also discovered there was a significant difference in the size of SRRM2 among all animal species, with lengths ranging from under 1000 amino acids to over 3500 amino acids. (Ilik et. al, 2020). They attributed this observation to the presence of an intrinsically disordered region, or “IDR,” where the number and order of amino acids could be altered without drastically impacting protein function. When the group analyzed related proteins, they found that the protein SON had a similar variation in length, suggesting that it also had an IDR present. (Ilik et. al, 2020) Previous work from Sharma et. al suggested that this protein may have a role to play in maintaining the organization of nuclear speckles because without SON protein present, splicing factors became disorganized (Sharma et. al, 2010). Given the similar variation in protein size present in SON and SRRM2, the group hypothesized that SRRM2 and SON could both be “essential” in the formation of speckles, (Ilik et. al, 2020).
To test their hypothesis, the group produced human HAP1 cells that lacked either SON or SRRM2 to determine the impact of the proteins on nuclear speckle development (Max-Planck-Geschellschaft, 2020). The group confirmed previous work suggesting that the removal of SON leads to more diffuse speckles. However, they also found that when SON production was inhibited in cells with severely “truncated” versions of SRRM2, the nuclear speckles dissolved completely into component proteins (Max-Planck-Geschellschaft, 2020). Similarly, the group found that when both SON and SRRM2 were completely inhibited in the HEK293 cell line, while other proteins present in speckles dispersed fully into the nucleoplasm (the fluid inside the nucleus) suggesting that both SRRM2 and SON are necessary for speckle formation (Ilik et. al, 2020). Given these results, contributing author Ibrahim Ilik stated that the group intends to further investigate how these proteins bind to molecules other than SC35 to determine other roles they may play in transcription (Max-Planck-Geschellschaft, 2020). Additionally, the discovery that SC35 binds primarily to SRRM2 over SRSF2 may necessitate further investigation into the role of nuclear speckles in various diseases. Co-author Tugce Aktaş also stated that this new role for SC35 may bring “entirely new perspectives” into the disease mechanisms for diseases like Huntington’s Disease and spinocerebellar ataxia (Max-Planck-Geschellschaft, 2020).
References
Clancy, S. (2008)RNA splicing: introns, exons and spliceosome. Nature Education 1(1):31
Fu, X., & Maniatis, T. (1990). Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus.Nature, 343(6257), 437-441. doi:10.1038/343437a0
Ilik, I. A., Malszycki, M., Lübke, A. K., Schade, C., Meierhofer, D., & Aktaş, T. (2020). SON and SRRM2 are essential for nuclear speckle formation.ELife, 9. doi:10.7554/elife.60579
Lamond, A. I., & Spector, D. L. (2003). Nuclear speckles: A model for nuclear organelles.Nature Reviews Molecular Cell Biology, 4(8), 605-612. doi:10.1038/nrm1172
Max-Planck-Gesellschaft. (2020, November 30). Getting to the core of nuclear speckles: Scaffold of sub-cellular structures identified after a hundred years.ScienceDaily. Retrieved December 22, 2020 from www.sciencedaily.com/releases/2020/11/201130113552.htm
Spector, D. L., & Lamond, A. I. (2011). Nuclear speckles.Cold Spring Harbor perspectives in biology, 3(2), a000646. https://doi.org/10.1101/cshperspect.a000646
Sharma, A., Takata, H., Shibahara, K., Bubulya, A., & Bubulya, P. A. (2010). Son Is Essential for Nuclear Speckle Organization and Cell Cycle Progression.Molecular Biology of the Cell, 21(4), 650-663. doi:10.1091/mbc.e09-02-0126
Related Posts
When Bacteria Band Together – How Interactions Between Bacteria Can Change the Course of Disease
Source: Adrian Lange Bacteria are constantly fighting to preserve themselves;...
Read MoreLike Owls, These Tiny Desert Dinosaurs Hunted in the Dark
Figure 1: Animated drawing of the extinct theropod Shuvuuia Source:...
Read MoreSingle Cell RNA Sequencing Enhances Understanding of Tumor Genetic Diversity
Figure 1: Researchers have shown that tumor cells are a...
Read MoreAndrew Sasser


Comments are closed.