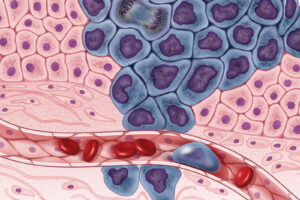
Figure: In a 2019 study, researchers used a microbial sample from the gut of an individual with the blood type AB+ to identify an enzyme combination that converts type A blood to type O, also known as the “universal donor” blood type. (Source: Wikimedia Commons, ICSident)
Many of us learned in high school biology class that type O blood is the “universal donor”: people with type O blood can only receive blood transfusions from other type O individuals, but people whose blood type is A, B, or AB can all receive type O blood. It is for this reason that type O blood is highly valuable and often in limited supply (Rahfeld & Withers, 2019). But what if there was a better way to ensure that a supply of universal donor blood is available to everyone who needs it?
Converting one blood type into another involves altering the antigens on the plasma membranes of red blood cells (RBCs). In all the ABO blood types, the RBC surface contains a short chain of sugar molecules called the H antigen (Rahfeld & Withers, 2019). In individuals with type O blood, only the H antigen is present. However, in people with blood types A and B, enzymes add the terminal carbohydrate chains GalNAc (for type A) or Gal (for type B) to the H antigen. Blood type AB means that both GalNAc and Gal are present on the RBC surface (Rahfeld et al., 2019). Using an enzyme to remove these terminal carbohydrates from the membranes of type A or B RBCs, while leaving the H antigen intact, would result in type O blood. If this could be done on a large scale, it could make more type O blood available for transfusion.
In a 2019 study, a team of researchers in Vancouver used metagenomic analysis, a technique that involves screening all the genomes contained within a microbial community or environmental sample, to search the gut microbiome of an individual with the blood type AB+ for microbial enzymes that might be able to convert type A or B blood into type O (Marchesi & Ravel, 2015; Rahfeld et al., 2019; Thomas, Gilbert, & Meyer, 2012). It might seem surprising that an enzyme that cleaves blood antigens would have evolved in intestinal bacteria. However, the authors chose to sample the human gut microbiome because the A and B antigens are also found in mucins, glycoproteins that line the intestinal wall to provide nourishment to bacteria and protection from invading microbes (Linden et al., 2008). Therefore, it would be expected that gut bacteria would express enzymes that also happen to remove the terminal sugars from RBCs (Rahfeld et al., 2019).
The researchers extracted microbial DNA from a fecal sample and isolated large fragments of the DNA that each contained multiple genes. They then inserted these bacterial DNA fragments into plasmids – small, circular, double-stranded bacterial DNA molecules found outside of a bacterium’s single chromosome (Shintani et al., 2015). In this study, the researchers packaged the plasmids into viral particles called bacteriophages, which they then used to inject the plasmids into Escherichia coli bacteria (Rahfeld et al., 2019). As these E. coli bacterial cells replicated, the plasmids were also replicated, resulting in a library of plasmids containing over 19,000 different clones of DNA fragments from the original bacteria (Rahfeld et al., 2019).
By screening the E. coli containing these plasmids with fluorescently labeled Gal and GalNAc substrates in the lab, the researchers identified eleven enzymes that cleaved the A and/or B antigens (Rahfeld et al., 2019). When used in combination, two of these enzymes, FpGalNAc deacetylase and FpGalNase, successfully removed the A antigen from the RBC surface under normal blood pH conditions. Previous techniques required a low pH that was more acidic than is optimal for RBCs or necessitated large quantities of enzyme that made the process impractical for routine usage (Rahfeld & Withers, 2019). While the researchers caution that further studies are necessary to determine whether the enzymes can remove all the A antigen from the RBC surface and to assess for any residual reactivity with human anti-A antibodies, the preliminary data appear promising, and the approach was able to change the type of a full unit of blood from A to O (Rahfeld et al., 2019).
If further research demonstrates the approach to be safe and effective, it could have implications well beyond blood transfusion. A recent study published in Science Translational Medicine tested the enzyme combination in type A human donor lungs, where it removed more than 97% of the A antigens from the blood vessel linings in 4 hours (Wang et al., 2022). Because the ABO antigen is expressed on most of the body’s cells and must typically be matched to the donor organ before a transplant can proceed, the conversion of lungs, kidneys, hearts, and other organs to type O would help ensure access to lifesaving organs for type O individuals, who are statistically more likely to die while awaiting a lung transplant than people with blood types A or B (Rahfeld & Withers, 2019; Wang et al., 2022). Perhaps, one day, donor organs will no longer need to come from someone with the same blood type as the recipient, and we can all be universal donors.
References
Linden, S. K., Sutton, P., Karlsson, N. G., Korolik, V., & McGuckin, M. A. (2008). Mucins in the mucosal barrier to infection. Mucosal Immunology 1(3), 183-197.
Marchesi, J. R., & Ravel, J. (2015). The vocabulary of microbiome research: A proposal. Microbiome, 3, 1-3.
Rahfeld, P., Sim, L., Moon, H., Constantinescu, I., Morgan-Lang, C., Hallam, S. J., Kizhakkedathu, J. N., & Withers, S. G. (2019). An enzymatic pathway in the human gut microbiome that converts A to universal O type blood. Nature Microbiology, 4, 1475-1485.
Rahfeld, P., & Withers, S. G. (2019). Toward universal donor blood: Enzymatic conversion of A and B to O type. Journal of Biological Chemistry 295(2), 325-334.
Shintani, M., Sanchez, Z. K., & Kimbara, K. (2015). Genomics of microbial plasmids: Classification and identification based on replication and transfer systems and host taxonomy. Frontiers in Microbiology, 6, 1-16.
Thomas, T., Gilbert, J., & Meyer, F. (2012). Metagenomics: A guide from sampling to data analysis. Microbial Informatics and Experimentation, 2(1), 3.
Wang, A., Ribeiro, R. V. P., Ali, A., Brambate, E., Abdelnour-Berchtold, E., Michaelsen, V., Zhang, Y., Rahfeld, P., Moon, H., Gokhale, H., Gazzalle, A., Pal, P., Liu, M., Waddell, T. K., Cserti-Gazdewich, C., Tinckam, K., Kizhakkedathu, J. N., West, L., Keshavjee, S.… Cypel, M. (2022). Ex vivo enzymatic treatment converts blood type A donor lungs into universal blood type lungs. Science Translational Medicine 14(632), 1-12.
Related Posts
Single Cell RNA Sequencing Enhances Understanding of Tumor Genetic Diversity
Figure 1: Researchers have shown that tumor cells are a...
Read MoreTransgender Health Disparities in Prisons
Transgender people are targets for victimization within the prison system...
Read MoreHow the Plague Changed Our Genes
Figure 1: An image of the Yersinia pestis bacterium that caused...
Read MoreA Spoonful of (Modified) Sugar as an Antiviral Medicine
Figure 1: A negative stain transmission electron micrograph of Influenza...
Read MoreDirecting Neural Stem Cells to Repair Damage
Figure 1: This depicts the ‘undifferentiated’ nature of pluripotent stem...
Read MoreConsidering the Microbiota-Gut-Brain-Axis in Cancer Research
Lead Author: Jillian Troth1 Co-Authors (alphabetical order): Roxanna Attar2, Julia Gainski3,...
Read MoreSofia Grant