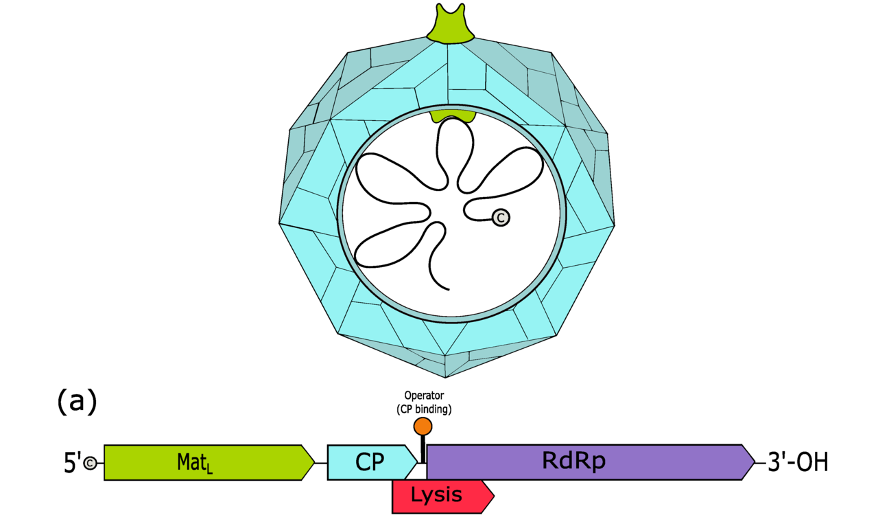
Figure 1: A computer-generated image of a bacteriophage called a levivirus along with its mapped genome. The Lysis gene is also known as sgi and produces antibiotic proteins
Source: Wikimedia Commons
We have a crisis on our hands: bacteria are developing resistance to our most effective antibiotics. Many of these are inhibitors of proteins that bacteria use to reproduce and cause infection. Due to overuse, the fact that many patients do not finish the full prescribed course of treatment, so-called antibiotic-resistant “superbugs” are developing mutant versions of their proteins that allow them to survive and cause harm even in the presence of medicine. Without discovering and/or developing new antibiotics, the possibility of a litany bacterial infections that we cannot treat, and a return to the dark ages before the first antibiotics were discovered, becomes very real (Ventola, 2015). Perhaps the best place to look for these life-saving drugs is within other infectious pathogens. Recently, researchers have turned to viruses.
It is not unusual for us to utilize other “dangerous” organisms for medicines. In fact, the first widely used antibiotic, Penicillin, was discovered by Alexander Fleming in 1928 when he noticed that the bacterium Staphylococcus aureus did not grow in plates contaminated with the mold Penicillium notatum. From that mold, he and other scientists work to develop the drug we know today as penicillin. Kardos and colleagues argue that Penicillin is the most important medicine ever created by humans due to the amount of lives it saved and how it inspired all antibiotics that followed. Today, penicillin is still being used against bacteria that have not developed resistance to it yet, including in cases of COPD, Syphilis, and Meningitis (Kardos, 2011). So, why not continue with this “enemy of my enemy” approach, and examine viruses for potential antibiotic molecules?
Researchers have been doing just this of late by examining the mechanisms of the viruses that infect bacteria, known as bacteriophages. Since the dawn of life, there has likely been an evolutionary battle between bacteria and the bacteriophages. This has led to a wide variety of families of bacteriophages, all with unique modes of infection and lysis (i.e., killing of host bacteria). While many of our medicines and own immune system struggle to fight off gram-negative bacteria (which possess an additional membrane that many drugs do not penetrate), bacteriophages face no such problem, making them an attractive resource for antibiotic activity (Brook, 2014).
Among these bacteriophages are leviviruses. Leviviruses are bacteriophages that have incredibly small genomes (on average, 3 genes) consisting of single-stranded RNA. Due to their tiny genomes, they are incredibly simple model viruses for study and researchers have been curious abou thow they use this small genome as effectively as they do to infect and kill bacteria. The gene products responsible for the lysis of bacteria infected by leviviruses are called Sgis (Chamakura, 2020).
Using 244 different Leviviruses, Chamakura and colleagues found 35 antibiotic Sgi proteins with activity against Escherichia coli, with some phages containing multiple such proteins. Additionally, many of these sgi genes were embedded as alternative reading frames (specifically, the +1 reading frame) within highly mutable regions of one of the protein coat gene. Essentially, it’s as if these viruses are hiding secret messages. If their genetic code is read one way, there are instructions to make the exterior of the viruses. However, if you started reading the instructions by reading from with the second letter instead of the first, the message explains how to make the protein responsible for lysis. Additionally, the words of these secret instructions are very susceptible to random change, altering the instructions to explain how to make an entirely new protein, one that may work in an entirely different way than the first.
The authors argue that if these phages are forced to evolve further Sgi proteins, they will likely develop a near-endless multitude of possible antibiotic candidates that we can use in the war against so-called “superbugs.” Then, the work will shift from being the responsibility of our microscopic allies to that of our own “soldiers” who will make these peptides usable in human bodies. This process will involve making the viral proteins consumable in an oral form, ensuring that they won’t cause any problems for human cells, and developing the correct dosage to ensure proper eradication of bacterial invaders.
The emergence of “pan-resistant” strains of bacteria (i.e., those that are resistant to many of our current antibiotics) is incredibly worrisome and is leading to a resurgence in incurable cases of bacterial pneumonia worldwide (Ventola, 2015). Perhaps the best course of action, like that taken in the battle against AIDS (a highly virus with an incredibly high rate of mutation and thus is quick to develop resistance to drugs), is to treat these strains with a “cocktail” of various drugs in an attempt to prevent mutation, as it is much harder to mutate a way to work around multiple inhibiting drugs. In order to develop these mixtures, we need more types of antibiotics that the bacteria are unable to counter (Brooks, 2014). The virus-sourced and human-altered antibiotics that can be developed from the leviviruses may serve that role, helping us eradicate old “superbugs” and new infectious bacteria alike.
References
Chamakura, K.R., Tran, J.S., O’Leary, C. et al. (2020). Rapid de novo evolution of lysis genes in single-stranded RNA phages. Nat Commun 11, 6009. https://doi.org/10.1038/s41467-020-19860-0
Kardos, N., Demain, A.L. (2011). Penicillin: the medicine with the greatest impact on therapeutic outcomes. Appl Microbiol Biotechnol 92, 677–687. https://doi.org/10.1007/s00253-011-3587-6
Texas A&M AgriLife Communications. (2021). ‘Hidden’ genes could be key in development of new antibiotics: Membrane-localized phage proteins may also help revitalize, enhance existing antibiotics. ScienceDaily. Retrieved February 8, 2021 from www.sciencedaily.com/releases/2021/02/210205121236.htm
Ventola C. L. (2015). The antibiotic resistance crisis: part 1: causes and threats. P & T : a peer-reviewed journal for formulary management, 40(4), 277–283.
Brooks B. A, Brooks A. E. (2014). Therapeutic strategies to combat antibiotic resistance, Advanced Drug Delivery Reviews 78, Pages 14-27, https://doi.org/10.1016/j.addr.2014.10.027.
Related Posts
The Immunological and Physiological Effects of SARS-COV-2 on the Human Body
Figure 1: The representation of cellular activity of SARS-COV-2 as...
Read MoreA New Perspective on Treating Asthma
Figure 1: This diagram displays the results of tissue remodeling...
Read MoreFrankie Carr