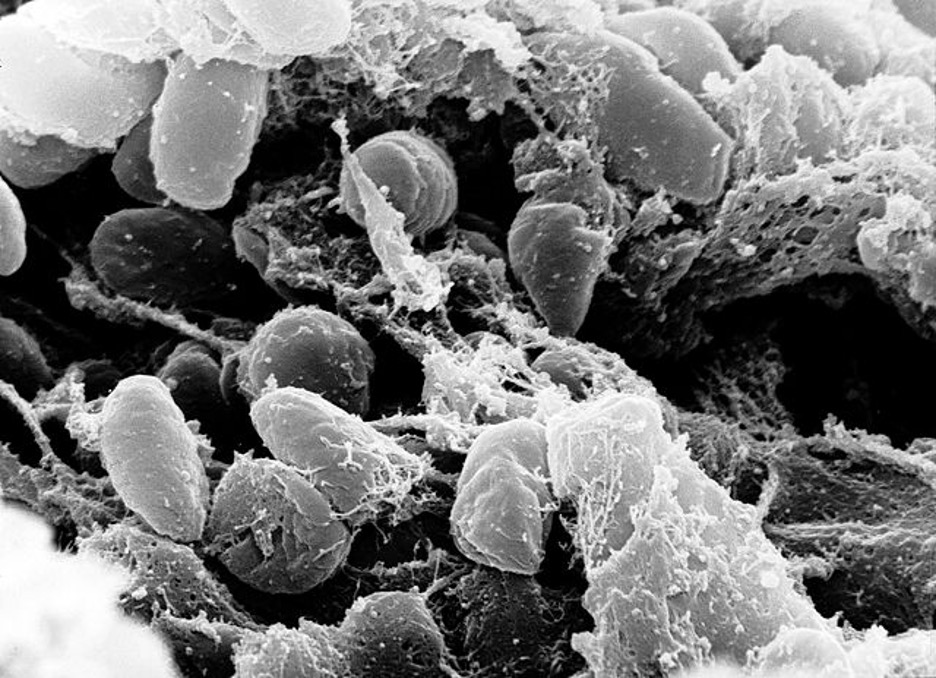
Figure 1: An image of the Yersinia pestis bacterium that caused the Black Plague. The image was taken with a scanning electron microscope using a sample collected from the digestive tract of an infected flea
Source: Wikimedia Commons, Rocky Mountain Laboratories
Some use the word “plague” to mean any infectious disease that infects many people, but the world originally referred to the highly infectious and dangerous disease caused by the bacterium Yersinia pestis, also known as the Black Plague, known for ravaging Europe during the 14th century. Y. pestis is a facultative intracellular pathogen, meaning that even if the bacterium is engulfed and swallowed by phagocytic immune cells, like macrophages, it can survive and reproduce. The Plague can present in three ways: Bubonic, Septicemic, or Pneumonic. Bubonic Plague is caused by the bacterium travelling from a wound (like a bite caused by an infected flea) to a lymph node, rapidly reproducing, and causing swelling there; this swollen lymph node is called a “bubo,” and swells up so much that it may be visible through the skin as a lump of swollen tissue. Sometimes, infection does not result in buboes or lung infection and the primary symptom is sepsis. Sepsis is a condition that occurs when the immune system’s response to pathogens in the bloodstream causes damage to the whole body, potentially resulting in organ failure. The lack of other tell-tale symptoms indicates the presence of Septicemic Plague (named due to the main symptom of this form being sepsis). Sebbane et al. showed that Septicemic Plague is more likely to occur if the bacteria do not enter the lymphatic system as the swelling of the lymph node is not possible, leaving sepsis as the predominant symptom. If the infection does not result in buboes but is present in the lungs, then it is called Pneumonic Plague. Such an infection can result in a bloody cough, potentially causing the bacterium to become an airborne pathogen that infects others in the immediate vicinity (Prentice & Rahalison, 2007; Sebbane et al., 2006).
Bubonic Plague is likely the most well-known form of the disease; it was responsible for the Black Death and spread throughout much of Afro Eurasia during the 14th century. Both fleas and rodents that were bitten and infected by the original fleas with the disease acted as vectors for the bacteria (Prentice & Rahalison, 2007). The Black Death was such a devastating epidemic that some estimates put its European death toll at around half of the population of the continent. While sporadic infections are still common, modern antibiotics have reduced the threat of the disease dramatically since the days of the Black Death (Immel et al., 2021).
It is not far-fetched to think that epidemic diseases may cause a genetic change in the human population via natural selection. Some people may possess alleles (a different version of a gene) in immunologically important genes that provide some sort of “advantage” – causing them to be better suited to fight off an infection or even prevent such an infection in the first place. These alleles and their advantages may cause their possessors to be more likely to survive the epidemic and thus be more likely to pass on those very alleles. In fact, such changes have been seen in response to cholera (Karlsson et al., 2013) and malaria (Kwiatkowski, 2005). Using this idea, a team of researchers recently undertook the task of comparing genetic samples from skeletons of those who seem to have died from the Bubonic Plague in the 16th century with modern citizens of the same small town in southern Germany to determine whether the disease has had any long-lasting effect on our bodies and genomes (Immel et al., 2021).
In comparing the genomes of the ancient denizens of the town with the modern ones, it was found that the two groups are genetically very similar with the exception of differential distributions of alleles in the FCN2 and NLRP14 genes along with the HLA complex of genes (Immel et al., 2021).
FCN2 produces the FCN2 protein, which is a Pattern Recognition Receptor (PRR). PRRs are proteins used by immune cells of the innate immune system to recognize and attack different kinds of pathogens; FCN2 is specifically used to recognize foreign, extracellular bacteria. The allelic changes in FCN2 include a change in the promoter (a sequence of nucleotides that is used to promote the transcription of the gene and thus production of the gene product) and one amino acid change. Using analytic techniques, the researchers determine that the FCN2 protein from the 16th century humans seems to have produced in lesser amounts (due to the promoter change) and had some form of differential binding, although the exact difference in binding is not known (Immel et al., 2021).
The gene product of NLRP14 is involved in the inflammasome complex, which are bacterial-specific PRRs that are used to recognize intracellular bacteria. So, when Y. pestis survives phagocytosis, NLRP14 signals to the immune cell that is infected intracellularly, triggering an immune response. The change in the gene from ancient to modern times is a change within the ligand-specificity domain of the protein, altering which molecules trigger the immunologic response. Thus, it is possible that the NLRP14 of those who died from the Plague did not bind to target molecules on the bacteria effectively, thus leading to their deaths (Immel et al., 2021).
Human Leukocyte Antigen (HLA) proteins are involved in presenting antigens (bits of proteins, fats, and sugars from within the cell or from extracellular material) to immune cells involved in the adaptive immune system (like virus and cancer-fighting Cytotoxic T Cells and immune-boosting T Helper Cells). HLA Class I proteins (encoded by HLA-A, HLA-B and HLA-C) are used to present molecules to Cytotoxic T Cells and HLA Class II proteins (encoded by HLA-DP, HLA-DQ, and HLA-DR) are used to present molecules to T Helper Cells. Different alleles of HLA genes produce molecules that are able to present different antigens. Thus, some people may have an HLA protein that can present antigens from Y. pestis (and other pathogens) while others may have HLA proteins that cannot. People with the HLA proteins that cannot present these antigens will be less likely to activate their adaptive immune system and thus less likely to fight off infection. The researchers found distinct allelic differences between the citizens of 16th century south Germany and their modern counterparts within the HLA-B, HLA-C, and HLA-DR genes. Therefore, the medieval Germans studied may have possessed HLAs that were less effective in Y. pestis antigen presentation, possibly contributing to their demise (Immel et al., 2021).
All of these alleles were likely present in the ancestors of the modern-day research participants, possibly (due to the combined or singular effects of the alleles) allowing them to survive the Black Death and thus to produce offspring bearing their genes. This is Darwin’s natural selection: those who possess traits that enable them to survive and/or produce more offspring will pass on those traits to their offspring. The Black Death took countless lives in medieval Europe, preventing many from passing on their genes. However, some people were able to survive (whether through fighting off infection or simply never getting infected), and thus gave their children the potentially life-saving alleles they possessed.
These findings have implications for the ongoing COVID-19 Pandemic (caused by the SARS-CoV-2 virus). As of May 2021, this disease has claimed at least 3,000,000 lives. However, around 160,000,000 people have been able to recover from infection and countless others (before vaccination) never got infected with the deadly virus (WHO, n.d.). It is likely that many of the survivors of this pandemic possess slight changes in immunologically important genes that helped them to survive, just as survivors of the black death likely possessed genetic variants that conferred the same sort of advantage. In surviving, they may pass those changes to their progeny, providing increased defense against the virus (and, hopefully, against related viruses), helping to shield future generations from the same desolation we have faced during the last year.
The human genome has been changed on a population level by malaria, cholera, and the Plague. Now, it will likely be changed by COVID-19.
References
Immel, A., Key, F. M., Szolek, A., Barquera, R., Robinson, M. K., Harrison, G. F., Palmer, W. H., Spyrou, M. A., Susat, J., Krause-Kyora, B., Bos, K. I., Forrest, S., Hernández-Zaragoza, D. I., Sauter, J., Solloch, U., Schmidt, A. H., Schuenemann, V. J., Reiter, E., Kairies, M. S., … Krause, J. (2021). Analysis of genomic DNA from medieval plague victims suggests long-term effect of Yersinia pestis on human immunity genes. Molecular Biology and Evolution, msab147. https://doi.org/10.1093/molbev/msab147
Karlsson, E. K., Harris, J. B., Tabrizi, S., Rahman, A., Shlyakhter, I., Patterson, N., O’Dushlaine, C., Schaffner, S. F., Gupta, S., Chowdhury, F., Sheikh, A., Shin, O. S., Ellis, C., Becker, C. E., Stuart, L. M., Calderwood, S. B., Ryan, E. T., Qadri, F., Sabeti, P. C., & LaRocque, R. C. (2013). Natural Selection in a Bangladeshi Population from the Cholera-Endemic Ganges River Delta. Science Translational Medicine, 5(192), 192ra86. https://doi.org/10.1126/scitranslmed.3006338
Kwiatkowski, D. P. (2005). How Malaria Has Affected the Human Genome and What Human Genetics Can Teach Us about Malaria. American Journal of Human Genetics, 77(2), 171–192.
Prentice, M. B., & Rahalison, L. (2007). Plague. The Lancet, 369(9568), 1196–1207. https://doi.org/10.1016/S0140-6736(07)60566-2
Sebbane, F., Jarrett, C. O., Gardner, D., Long, D., & Hinnebusch, B. J. (2006). Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proceedings of the National Academy of Sciences of the United States of America, 103(14), 5526–5530. https://doi.org/10.1073/pnas.0509544103
WHO. (n.d.). WHO Coronavirus (COVID-19) Dashboard. Retrieved May 21, 2021, from https://covid19.who.int
Related Posts
Intergenerational Trauma: How a history of pain brings biological costs
Source: Nicolas Raymond, CC BY 2.0 A few years ago,...
Read MoreUnderstanding the Social Factors Affecting Cancer Therapy
Cover Image: A patient being prepared for radiation therapy. (Source:...
Read MoreThe Effects of Friends and Family on Daily Stress
Figure 1: This figure shows several of the different effects...
Read MoreFrankie Carr