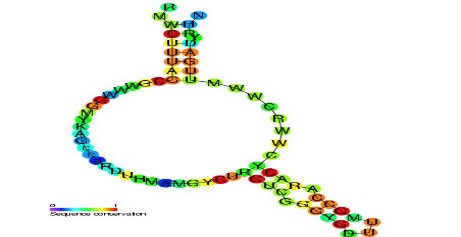
Figure 1: Predicted secondary structure and sequence of a retron msr RNA with a sequence conservation metric that compares phylogenetic sequence alignments between species
Source: Wikimedia Commons
The gene editing tool CRISPR-Cas9 is considered to be one of the most significant inventions of the 21st century. Its developers, Jennifer Doudna and Emmanuelle Charpentier, were awarded the 2020 Nobel Prize in chemistry. In line with the Nobel foundation’s philosophy of “for the greatest benefit to humankind,” this tool promises an avenue to break free from the confines of maladaptive genes. However, lurking in the shadows of CRISPR, an old but less well-known tool, the retron, is gaining ground as a tool to augment the diversity of the scientist’s gene editing repertoire.
Retrons are bacterial genetic elements composed of a reverse transcriptase (an enzyme that can synthesize DNA from RNA) and a non-coding RNA. The first retron was documented in 1987 when Inouye et al. discovered a short multi-copy single stranded DNA (MsDNA) in the cell of the bacteria Myxococcus xanthus. MsDNA consists of a chimeric RNA/DNA molecule in which the RNA and DNA components are covalently linked to form a stem loop sequence. The coding region of the MsDNA is the retron which encodes a reverse transcriptase essential for MsDNA synthesis (Inouye, 1987). Despite close to 30 years of research, the biological function of the retron and its mechanisms of genetic transmission have remained enigmatic.
Over the years, the retron has been found to play a role in a diverse set of E.coli biochemical pathways, active when the bacterium is starved of phosphate, and also contributing to pathogenesis and cell specialization (Elfenbein, 2015; Herzer 1996; Simon, 2019). In a more recent paper, however, Millman et al. conclusively classified the retron to have evolved phylogenetically, just like CRISPR, as an immunologic mechanism targeting phages (viruses that infect bacteria.)
Recent advances have enabled the integration of CRISPR and retron systems to create “next generation” gene editors such as Cas9 Retron precISe Parallel Editing via homologY (CRISPEY). Unlike CRISPR which has a limited scope and can only introduce a limited number of edits, CRISPEY can introduce thousands of specific genetic edits in a single experiment. The wider scope of the CRISPEY system stems from the retron’s reverse transcriptase subunit which allows continuous synthesis of DNA sequences. Recently CRISPEY, developed by researchers at Stanford, was used to produce mutant yeast genomes that enabled identification of key genes required for yeast to thrive in a glucose medium (Pennisi, 2020). Currently, however, CRISPEY can only be used to edit single cells, so there is a lot of work to be done before it is ready for prime time as a gene editing tool.
What makes retrons indispensable in future biotechnological endeavors? Once harnessed and improved, may serve as “in vivo” sources of donor DNA without the need to introduce external oligonucleotide sequences via electroporation or transfection with viral vectors. (Lundstrom, 2018). Further optimization in the delivery of diverse mutations in multicellular settings may prove useful in generating highly programmable transgenic organisms that may accelerate biotechnology research. The recent renaissance in retron research may fuel yet more novel applications.
References
Dhundale, A., Lampson, B., Furuichi, T., Inouye, M., & Inouye, S. (1987). Structure of MsDNA from Myxococcus xanthus: evidence for a long, self-annealing RNA precursor for the covalently linked branched RNA. Cell, 51(6), 1105-1112.
Elfenbein, J. R., Knodler, L. A., Nakayasu, E. S., Ansong, C., Brewer, H. M., Bogomolnaya, L., … & Andrews-Polymenis, H. L. (2015). Multicopy single-stranded DNA directs intestinal colonization of enteric pathogens. PLoS genetics, 11(9), e1005472.
Herzer, P. J. (1996). Starvation-induced expression of retron-Ec107 and the role of ppGpp in multicopy single-stranded DNA production. Journal of bacteriology, 178(15), 4438-4444.
Lundstrom, K. (2018). Viral vectors in gene therapy. Diseases, 6(2), 42. https://doi.org/10.3390/diseases6020042
Millman, A., Bernheim, A., Stokar-Avihail, A., Fedorenko, T., Vochek, M., Leavitt, A., & Sorek, R. (2020). Bacterial retrons function in anti-phage defense. Cell.183 (6), 1551-1561.
Nobel Media AB (2020). Retrieved December 22, 2020 from https://www.nobelprize.org/prizes/chemistry/2020/press-release/
Pennisi, E. (2020). Like CRISPR, mystery gene editor began as a virus fighter. https://science.sciencemag.org/content/370/6519/898.summary
Simon, A. J., Ellington, A. D., & Finkelstein, I. J. (2019). Retrons and their applications in genome engineering. Nucleic acids research, 47(21), 11007-11019. https://academic.oup.com/nar/article/47/21/11007/5584520
Related Posts
Whale Size Correlates with Feeding Pattern Efficiency
Figure 1: Blue Whale Surfacing off the Coast of Oregon...
Read MoreModeling Maximum Running Speed in Animals
Figure: Two cheetahs lie side by side. As the fastest...
Read MoreA New Perspective on Treating Asthma
Figure 1: This diagram displays the results of tissue remodeling...
Read MoreWhitney Nicanor Mabwi