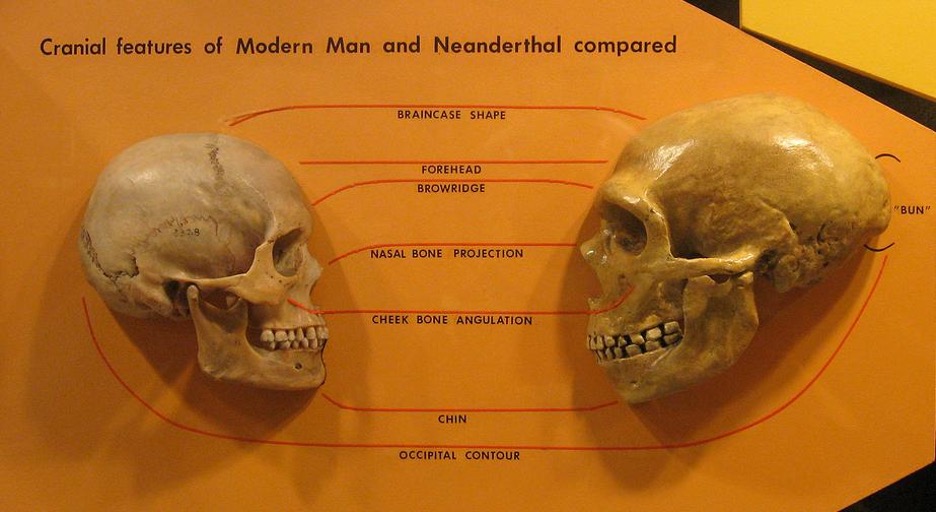
Figure 1: A comparison of the skulls of a human (left) and Neanderthal (right).
Source: Wikimedia Commons
Studies of evolution rely heavily on the use of fossils. However, in order to understand the differences in intelligence between humans and one of our closest relatives, the Neanderthals, it is necessary to study the differences in the brains of these species. Unfortunately, the brain’s fatty structure does not lend itself well to fossilization, so scientists have nothing to go off of — nothing, but DNA.
After analyzing the genomes of collected fossils of Neanderthals, researchers at University of California San Diego were able to use the differences in the protein-coding regions between the DNA of specimens of ancient Homo sapiens and Homo neanderthalensis to try to determine which changes could have an effect on brain development, structure, and function that were present during the time that both species roamed and thought on this planet. Changes between H. neanderthalensis and modern humans may reflect the role of the natural selection that has occurred since their extinction and thus were not considered. One such gene, NOVA1, encodes a protein that acts as a regulator of alternative splicing (a pathway used to generate multiple protein forms from a given gene) that is used during the development of the nervous system and for the formation of synapses (the gaps between neurons through which they communicate) (Trujillo et al., 2021).
The only nonsynonymous change (i.e., the only change that actually results in a change in protein sequence) in NOVA1 between the two species is a small one: I200V. This change, from isoleucine to valine, affected one of the three domains of the protein responsible for binding to target mRNA molecules and thus altered the expression of the mRNA that will eventually be transcribed into proteins that affect nervous system and synapse development. Based on this, the researchers theorized this change would play a role in generating differential activity of the protein between the two species (Trujilo et al., 2021).
To test this, the team used CRISPR-Cas9 to edit the genome of a line of pluripotent human stem cells (cells that can differentiate into any kind of cell that is present in the body). This resulted in cells that have the isoleucine in its NOVA1 protein at the 200th position, essentially giving them the Neanderthal version of the protein. The team then grew these cells (as well as cells with the valine containing NOVA1 that was seen in ancient humans) in an environment that encouraged them to develop in nervous system tissue, generating brain “organoids” (Trujilo et al., 2021).
These organoids are simple, smaller models of organs, though they are isolated and not connected to other systems (nervous, vascular, etc.) like organs within a body are. Organoids are a relatively new tool that scientists can use to study the development and growth of organs during embryonic development and to model the effects of genetic diseases, infections, and cancer on organs (Lancaster & Knoblich, 2014). The researchers from UCSD used this technique to develop “brains in a jar” of both ancient humans and Neanderthals in order to see how they might look and act differently.
Once the researchers grew these “brains,” they examined their structures and protein products. The two forms of NOVA1 had differential binding preferences, which led to them producing different versions of mRNA transcripts of various target genes. This manifested itself in an overall decrease in proteins on either side of synapses of the nerves in the Neanderthal “brain” when compared to that of the human. When testing how this might impact the firing of these nerves, the researchers noted that the Neanderthal organoid fired more rapidly, with more variability, and in a less synchronized way when compared to the firing of the human organoid (Trujilo et al., 2021). The researchers concluded that this pattern of firing resembled a more primitive firing pattern, such as that seen in newborn individuals of non-human primates (ScienceDaily).
There are certainly limitations in the study, such as the fact that these are not full brains (they lack the additional scaffolding of bones, skin, blood vessels, etc. and that might be contributing to these changes) and that these organoids were produced using gene-editing technology (meaning that there may be some off-target effects). Despite these issues, the results of the study are astounding. One simple change resulted in differential brain structure and firing. Perhaps the other changes present in the Neanderthal genome might counteract these changes, or they might exaggerate them further. The research team at USCD plans to examine just how the nonsynonymous differences in the other 60 genes they identified may affect brain development. This may provide insight into the differences between our brains and our ancestral cousins. Perhaps these differences led to more organized, smarter brains that allowed us to outlast the Neanderthals and continue to thrive today. In fact, they may have made our brains sophisticated enough to eventually do what we are doing now: studying those very differences.
References
Lancaster, M. A., & Knoblich, J. A. (2014). Organogenesis in a dish: Modeling development and disease using organoid technologies. Science, 345(6194). https://doi.org/10.1126/science.1247125
Trujillo, C. A., Rice, E. S., Schaefer, N. K., Chaim, I. A., Wheeler, E. C., Madrigal, A. A., Buchanan, J., Preissl, S., Wang, A., Negraes, P. D., Szeto, R. A., Herai, R. H., Huseynov, A., Ferraz, M. S. A., Borges, F. S., Kihara, A. H., Byrne, A., Marin, M., Vollmers, C., … Muotri, A. R. (2021). Reintroduction of the archaic variant of NOVA1 in cortical organoids alters neurodevelopment. Science, 371(6530). https://doi.org/10.1126/science.aax2537
University of California – San Diego. (2021, February 11). How a single gene alteration may have separated modern humans from predecessors: Novel study used brain organoids genetically modified to mimic now-extinct Neanderthals. ScienceDaily. Retrieved February 15, 2021 from www.sciencedaily.com/releases/2021/02/210211144418.html
Related Posts
Regulating Regulatory T Cells to Fight Cancer
Figure 1: The different ways a tumor can provide protection...
Read MoreBispecific Antibody Recruits Vγ9+ γδ T cells for Leukemia Treatment
Figure 1: The image is taken from an elderly woman...
Read MoreGenetic Cheating? Identifying CRISPR/Cas Gene Doping
Figure 1: Lance Armstrong, a well-known professional cyclist, has been...
Read MoreFrankie Carr